Understanding ARTS results
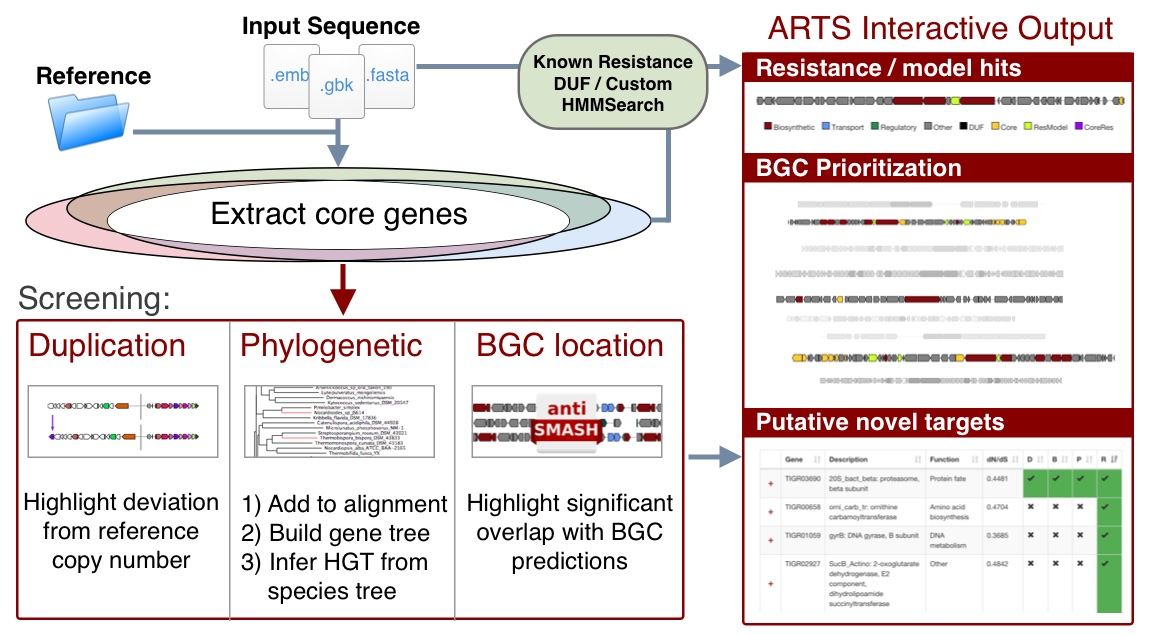
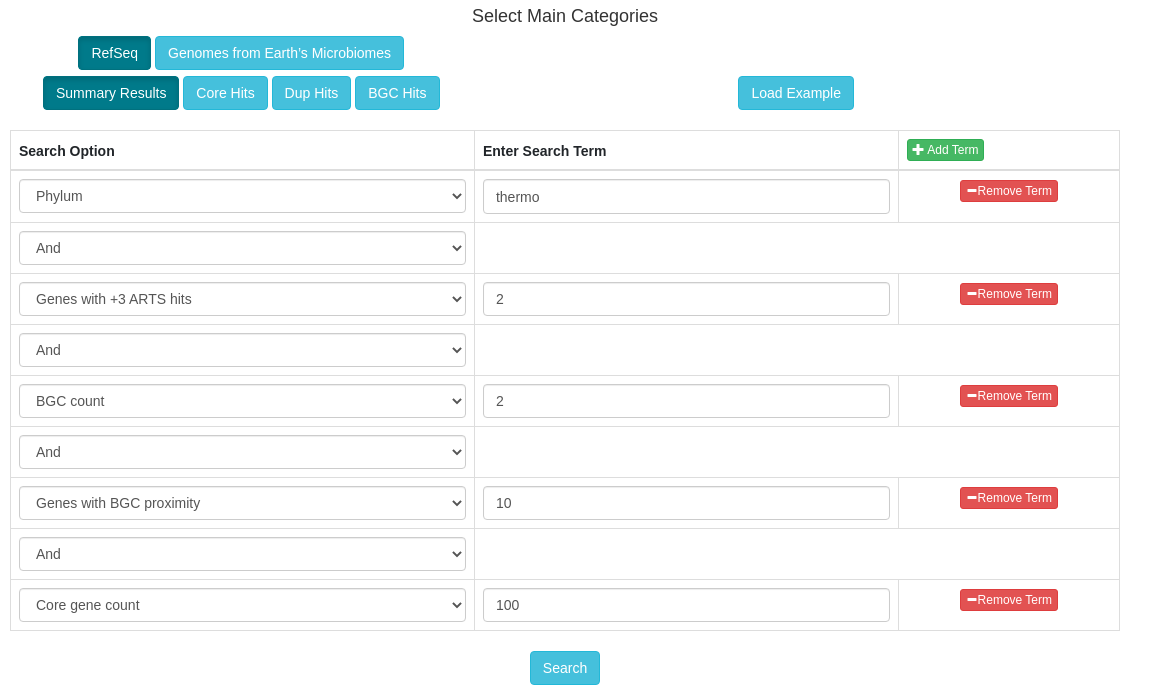
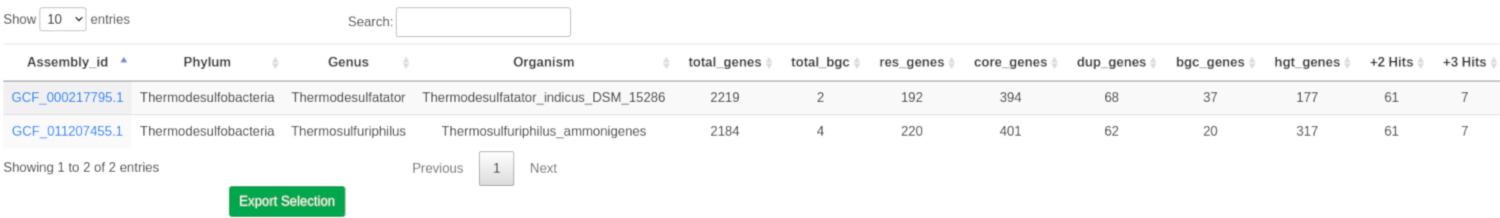
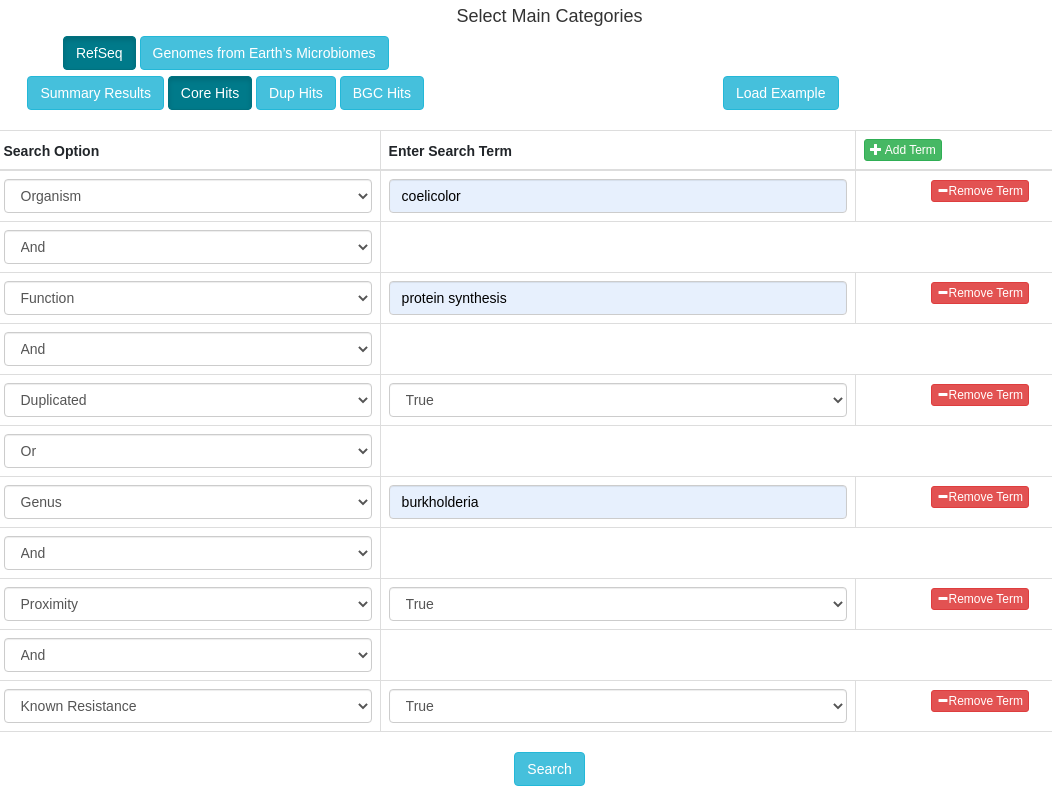
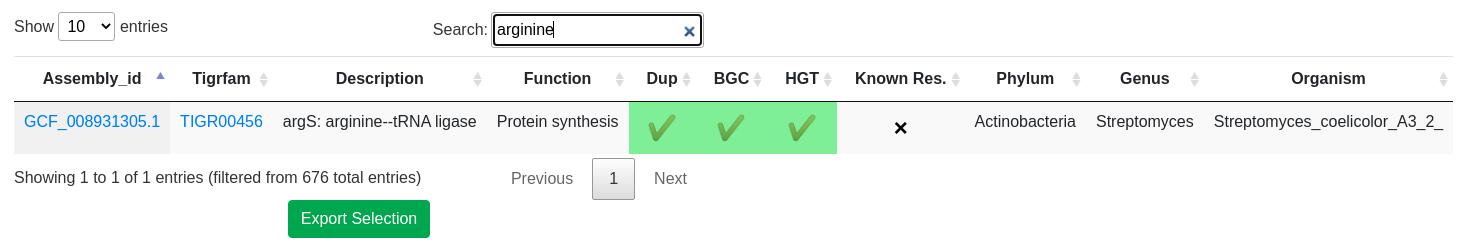
The goals of this tool are to automate the process of performing target direct genome mining (TDGM); search for potential novel antibiotic targets, and prioritize putative secondary metabolite gene clusters.
There are three fundamental ARTS criteria for TDGM as listed below:
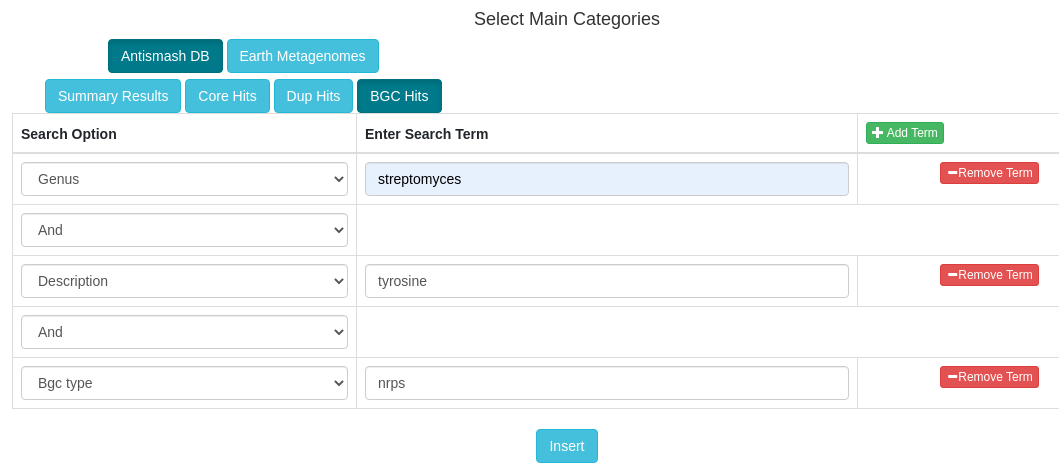
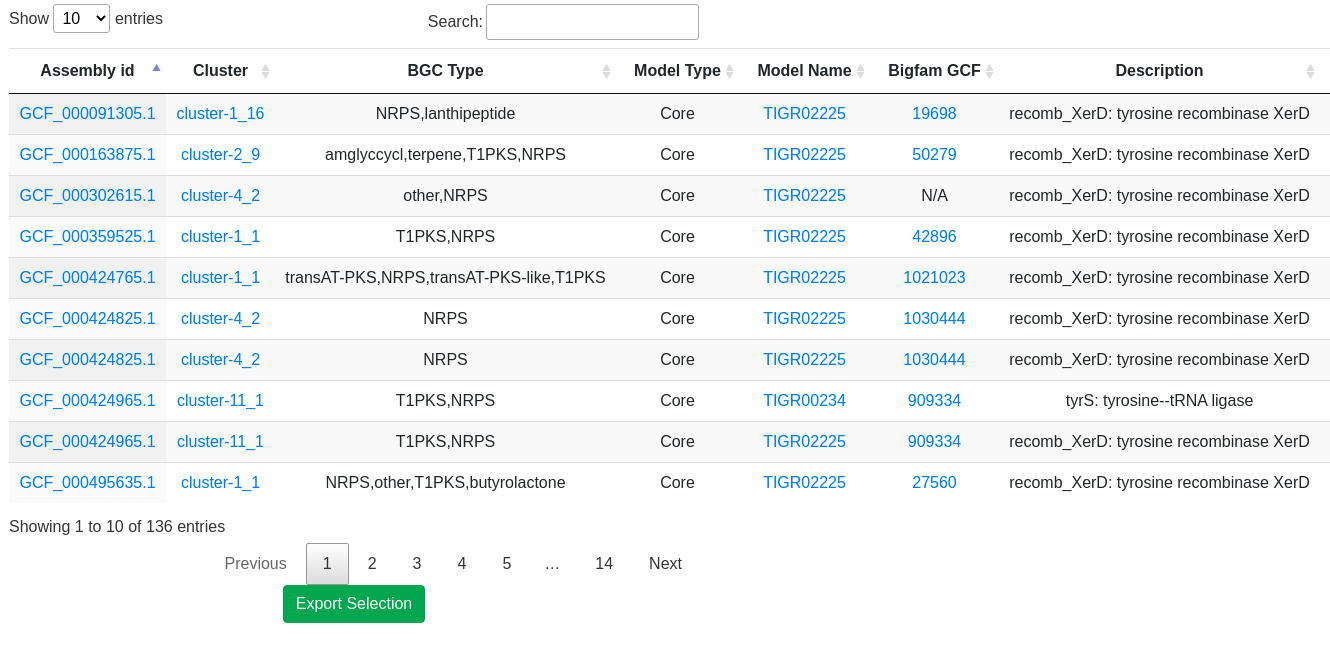
- Proximity check: Cross reference locations with Secondary Metabolite gene clusters. For more information about BGC prediction, please visit antiSMASH help page
- Uncommon duplication check: Highlight potential repurposed primary metabolism genes
- Phylogenetic incongruence: Highlight essential genes with evidence of inter-genus horizontal transfer
Please visit ARTS help page for more information.